|
Table of Contents |
|
1. Introduction
2. Thin Plate Splines: Tps 3. Spatial Process Models: Krig 4. Simulating Random Fields (sim.rf) 5. Spatial Predictions for Large Data Sets
|
|
Printable version of Fields Manual
Back to Fields Home Page |
Data obtained from Gomez and Hazen (1970, Tables 19 and 20) on coal ash for the Robena Mine Property in Greene County Pennsylvania.
Overall plot of the data.
# -- Create an image plot of the data. For fun, add the contour lines.
look <- as.image( coalash$y, x=coalash$x)
image.plot( look)
contour( look, add=T, labex=0)

# -- Perform a Krig fit using exponential covariance function with range
# -- paramter, theta=2.5 (choice of theta will be explained below).
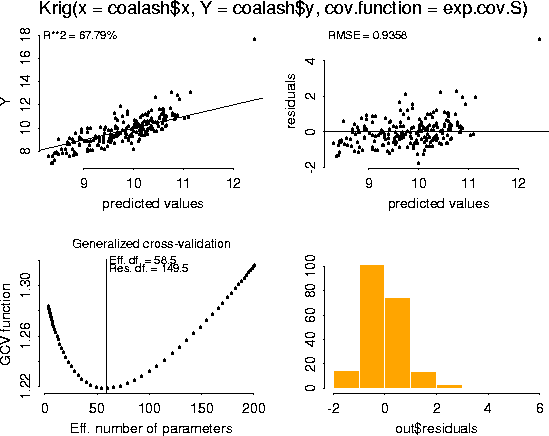
fit <- Krig( coalash$x, coalash$y, cov.function = exp.cov.S, theta=2.5)
summary( fit)
Call:
Krig(x = coalash$x, Y = coalash$y, cov.function = exp.cov.S)
Number of Observations: 208 Number of unique points: 208 Degree of polynomial null space ( base model): 1 Number of parameters in the null space 3 Effective degrees of freedom: 29.7 Residual degrees of freedom: 178.3 MLE sigma 1.02 GCV est. sigma 1.018 MLE rho 0.2829 Scale used for covariance (rho) 0.2829 Scale used for nugget (sigma^2) 1.04 lambda (sigma2/rho) 3.675 RESIDUAL SUMMARY: min 1st Q median 3rd Q max -2.169 -0.6578 -0.09917 0.4115 6.169 COVARIANCE MODEL: exp.cov.S DETAILS ON SMOOTHING PARAMETER: Method used: GCV Cost: 1 lambda trA GCV GCV.one GCV.model shat 3.675 29.69 1.209 1.209 NA 1.018 Summary of estimates for lambda lambda trA GCV shat GCV 3.675 29.69 1.209 1.018 GCV.one 3.675 29.69 1.209 1.018
plot( fit)
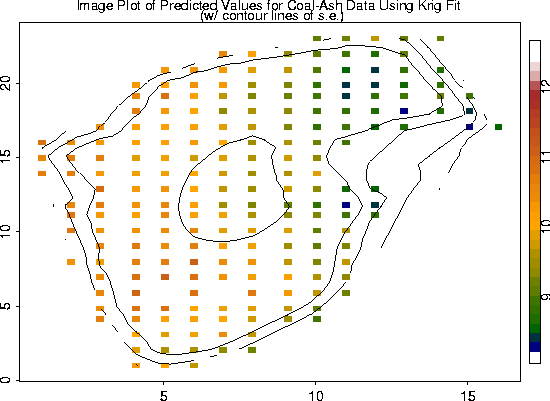
# -- We will use the S-Plus generic function, predict, to make predictions,
# -- using the Krig object fit.
# -- We also added the contours of the standard errors of the estimates.
fit.pred <- predict( fit)
look <- as.image( fit.pred, x=coalash$x)
image.plot( look)
fit.se <- predict.se( fit)
look2 <- as.image (fit.se, x=coalash$x)
contour( look2, add=T, labex=0)
| theta | GCV Minimum |
0.5 0.75 1.0 1.25 1.5 1.75 2.0 2.25 2.5 2.75 3.0 3.25 3.5 3.75 4.0 4.25 5.0 10.0 100.0 |
1.225 1.221 1.218 1.216 1.214 1.212 1.211 1.21 1.209 1.209 1.208 1.208 1.207 1.207 1.207 1.207 1.206 1.205 1.205 |
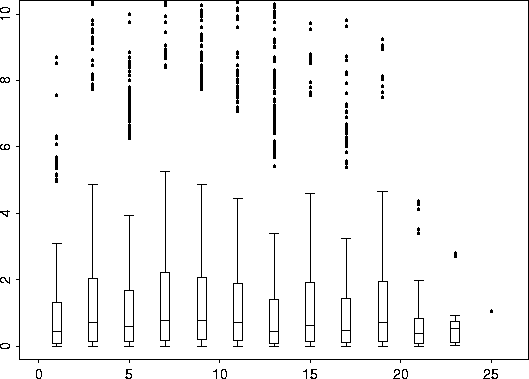
coalash.vgram <- vgram( coalash$x, coalash$y)
# -- coalash.vgram is a list object. Where "d" are the distances,
# -- coalash.vgram$vgram is the variogram, and call tells how the vgram function was called.
names( coalash.vgram)
[1] "d" "vgram" "call"
bplot.xy( coalash.vgram$d, coalash.vgram$vgram)
 |
 |
Now we use the S-Plus function, nls (Non Linear Least Squares Regression) to search for a suitable range parameter. Simultaneously, we search for possible values for sigma2 and rho which we can subsequently fix, if desired, for the Krig fit. vgram.fit <- nls( vgram ~ sigma2*(1-rho*exp(-d/theta)), coalash.vgram, start=list( sigma2= 9, rho=0.95, theta=1)) summary( vgram.fit)
Formula: vgram ~ sigma2 * (1 - rho * exp( - d/theta))
Parameters:
Value Std. Error t value
sigma2 1.696190 0.0334569 50.69770
rho 0.750553 0.2807990 2.67292
theta 1.635050 0.5580010 2.93019
Residual standard error: 3.60828 on 21523 degrees of freedom
Correlation of Parameter Estimates:
sigma2 rho
rho -0.346
theta 0.566 -0.877
fit <- Krig( coalash$x, coalash$y, theta=1.635050, sigma2=1.696190, rho=0.750553)
summary( fit)
CALL:
Krig(x = coalash$x, Y = coalash$y, rho = 0.750553, sigma2 = 1.69619, theta =
1.63505)
Number of Observations: 208
Number of unique points: 208
Degree of polynomial null space ( base model): 1
Number of parameters in the null space 3
Effective degrees of freedom: 48.8
Residual degrees of freedom: 159.2
MLE sigma 0.9604
GCV est. sigma 0.9647
MLE rho 0.4082
Scale used for covariance (rho) 0.7506
Scale used for nugget (sigma^2) 1.696
lambda (sigma2/rho) 2.26
RESIDUAL SUMMARY:
min 1st Q median 3rd Q max
-1.899 -0.5541 -0.1009 0.3808 5.486
COVARIANCE MODEL: exp.cov
DETAILS ON SMOOTHING PARAMETER:
Method used: user Cost: 1
lambda trA GCV GCV.one GCV.model shat
2.26 48.8 1.216 1.216 NA 0.9647
Summary of estimates for lambda
lambda trA GCV shat
GCV 3.422 37.27 1.213 0.9979
GCV.one 3.422 37.27 1.213 0.9979